Spectroscopy datasets often consist of high-dimensional measurements across different wavelengths or wavenumbers, structured in wide-format tables where each column represents a sample and each row corresponds to a spectral position. The tidyspec package provides a tidyverse-friendly toolkit designed to streamline the processing, visualization, and analysis of such spectral data in R.
By building on top of the dplyr,
ggplot2, and tidyr ecosystems,
tidyspec simplifies common workflows such as baseline
correction, unit conversion, normalization, filtering, and principal
component analysis (PCA). It enforces consistent treatment of spectral
dimensions using a customizable reference column (typically wavenumber
or wavelength), which can be easily set using
set_spec_wn().
This vignette demonstrates a typical usage of the package, from importing and visualizing spectral data to performing PCA and interpreting results. The functions included are designed to make spectral analysis in R more transparent, reproducible, and user-friendly for both beginners and advanced users.
Package overview
The package organizes utilities into 6 families, all prefixed with
spec_:
1. Transformationspec_abs2trans(): Convert absorbance to transmittance.
spec_trans2abs(): Convert transmittance to absorbance.
2. Normalizationspec_norm_01(): Normalize spectra to range [0, 1].
spec_norm_minmax(): Normalize to a custom range.
spec_norm_var(): Scale to standard deviation = 1.
3. Baseline Correctionspec_blc_rollingBall(): Correct baseline using rolling ball
algorithm. spec_blc_irls(): Correct using Iterative
Restricted Least Squares. spec_bl_*(): Return baseline
vectors (e.g., spec_bl_rollingBall).
4. Smoothingspec_smooth_avg(): Smooth with moving average.
spec_smooth_sga(): Smooth using Savitzky-Golay.
5. Derivativespec_diff(): Compute spectral derivatives.
6. Preview & I/Ospec_smartplot(): Static preview of spectra.
spec_smartplotly(): Interactive preview
({Plotly}). spec_read(): Import from .csv,
.txt, .xlsx, etc.
Data
The CoHAspec dataset is a spectral table in wide format,
where:
Rows: Wavenumbers (in cm⁻¹).
Columns: Samples (CoHA01, CoHA025, CoHA05, CoHA100).
Values: Absorbance/intensity measurements.
Here’s a preview of the data:
CoHAspec
#> # A tibble: 1,868 × 5
#> Wavenumber CoHA01 CoHA025 CoHA05 CoHA100
#> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 399. 0.871 1.36 1.17 1.05
#> 2 401. 0.893 1.24 1.05 0.925
#> 3 403. 0.910 1.20 0.997 0.876
#> 4 405. 0.914 1.19 0.982 0.867
#> 5 407. 0.908 1.18 0.965 0.857
#> 6 409. 0.887 1.14 0.936 0.828
#> 7 411. 0.856 1.08 0.902 0.791
#> 8 413. 0.819 1.03 0.865 0.748
#> 9 415. 0.789 0.988 0.838 0.717
#> 10 417. 0.768 1.000 0.860 0.735
#> # ℹ 1,858 more rowsWavenumber Handling
The function set_spec_wn simplifies the use of functions
by globally defining the column that contains the wave numbers. User can
check the wavenumber column with check_wn_col.
set_spec_wn("Wavenumber")
#> Successfully set 'Wavenumber' as the default wavenumber column.
check_wn_col()
#> The current wavenumber column is: WavenumberVisualize the data
Scientists who work with spectroscopy data always want to visualize
it after each operation. That’s why we created the
spec_smartplot and spec_smartplotly functions
to help users.
spec_smartplot creates a static visualization of the
spectra, while spec_smartplotly creates an interactive
visualization, allowing the user to navigate through the values of the
different data.
spec_smartplot(CoHAspec)
#> Warning: wn_col not specified. Using default value: Wavenumber.
#> This message is shown at most once every 2 hours.
#> Warning: xmin not specified. Using default value: 399.1992.
#> This message is shown at most once every 2 hours.
#> Warning: xmax not specified. Using default value: 3999.706.
#> This message is shown at most once every 2 hours.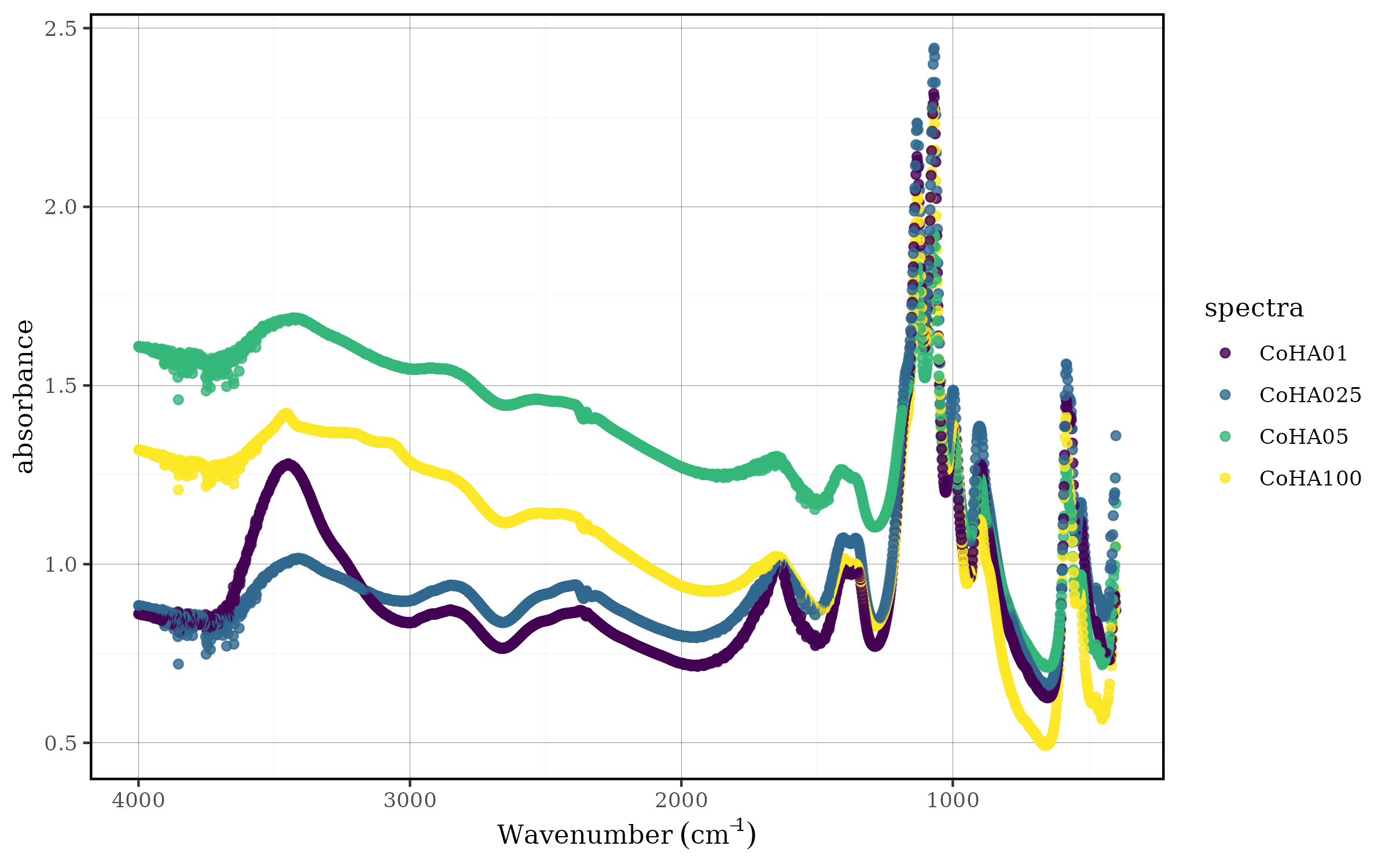
Static spectral plot showing all samples
spec_smartplotly(CoHAspec,geom = "line")These functions simplify the process by automating it, but we invite
users to make their own figures for presentations and publications. As
part of the efforts to create a community of tidyspec
users, we plan to create a ggplot2 extension for high-level
spectroscopy graphics. For now, you can produce ggplot2
graphs by pivoting the data:
CoHAspec %>%
tidyr::pivot_longer(cols = -Wavenumber,
names_to = "spectrums",
values_to = "absorbance") %>%
ggplot(aes(x = Wavenumber, y = absorbance, col = spectrums)) +
geom_line() #<------ Customize your data from here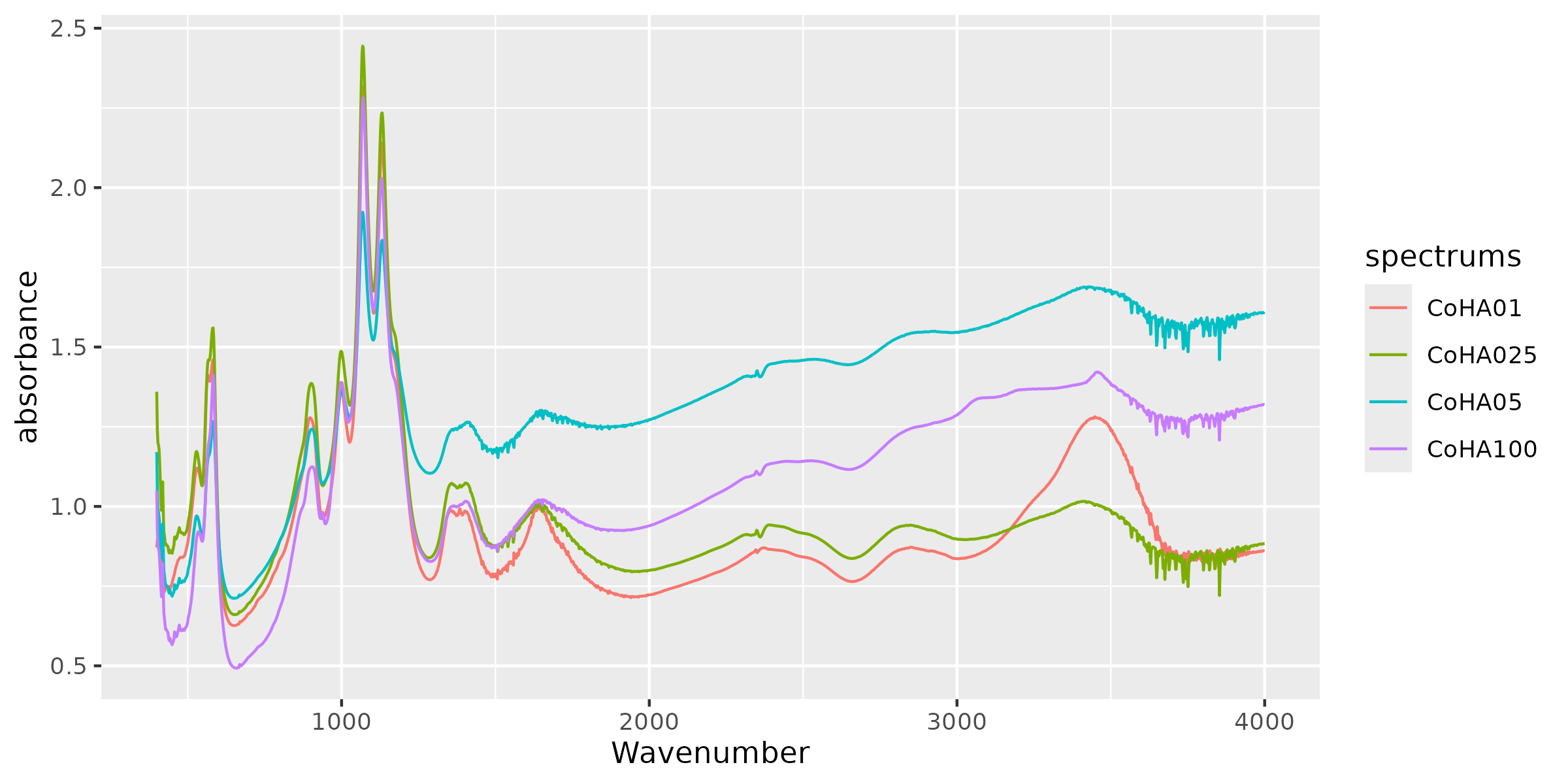
Custom ggplot2 visualization of spectral data
Convert to transmittance (and back to absorbance)
The functions spec_abs2trans and
spec_trans2abs are designed to easily convert the
spectras.
CoHAspec %>%
spec_abs2trans() %>%
spec_smartplot()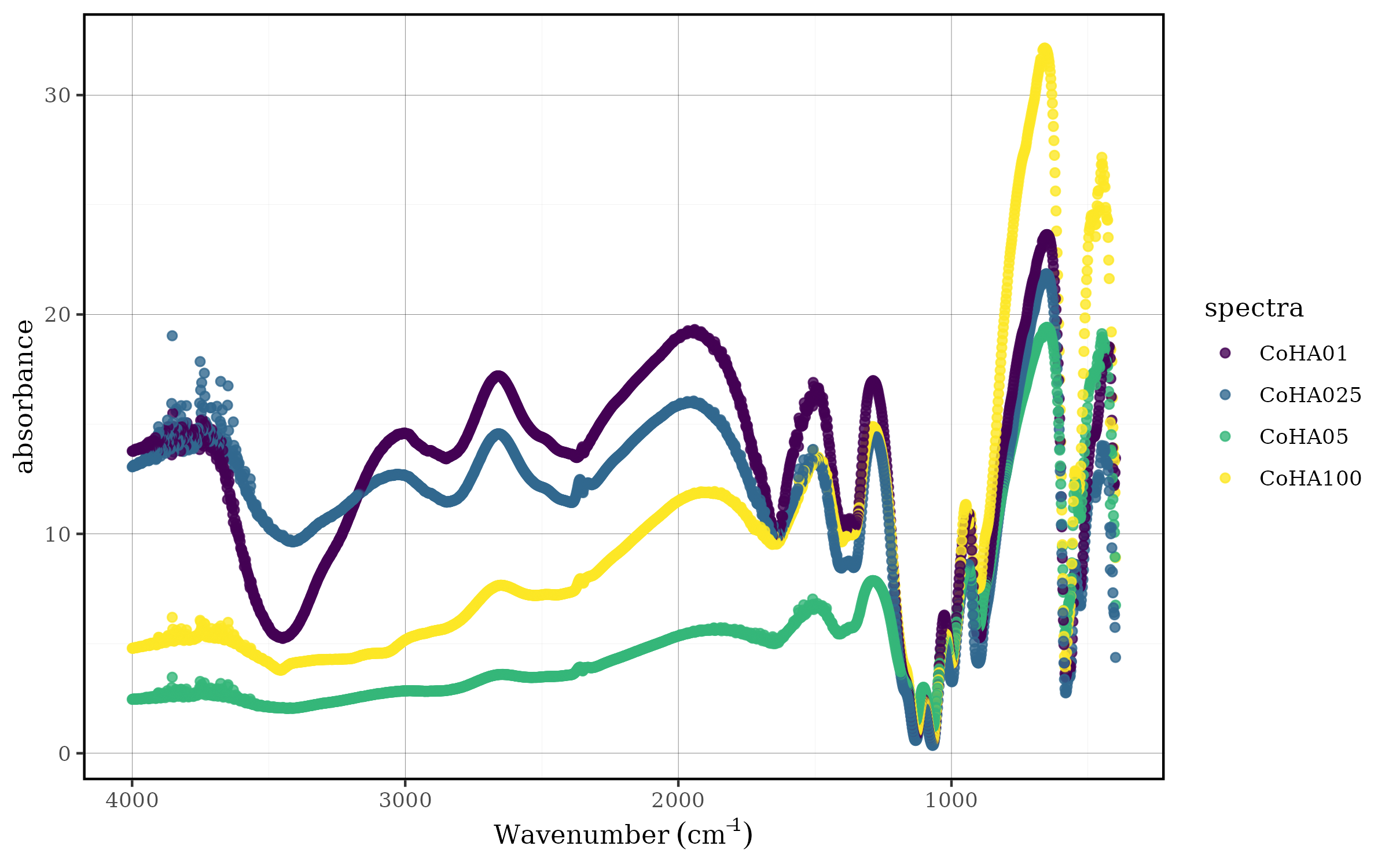
CoHAspec %>%
spec_abs2trans() %>%
spec_trans2abs() %>%
spec_smartplot()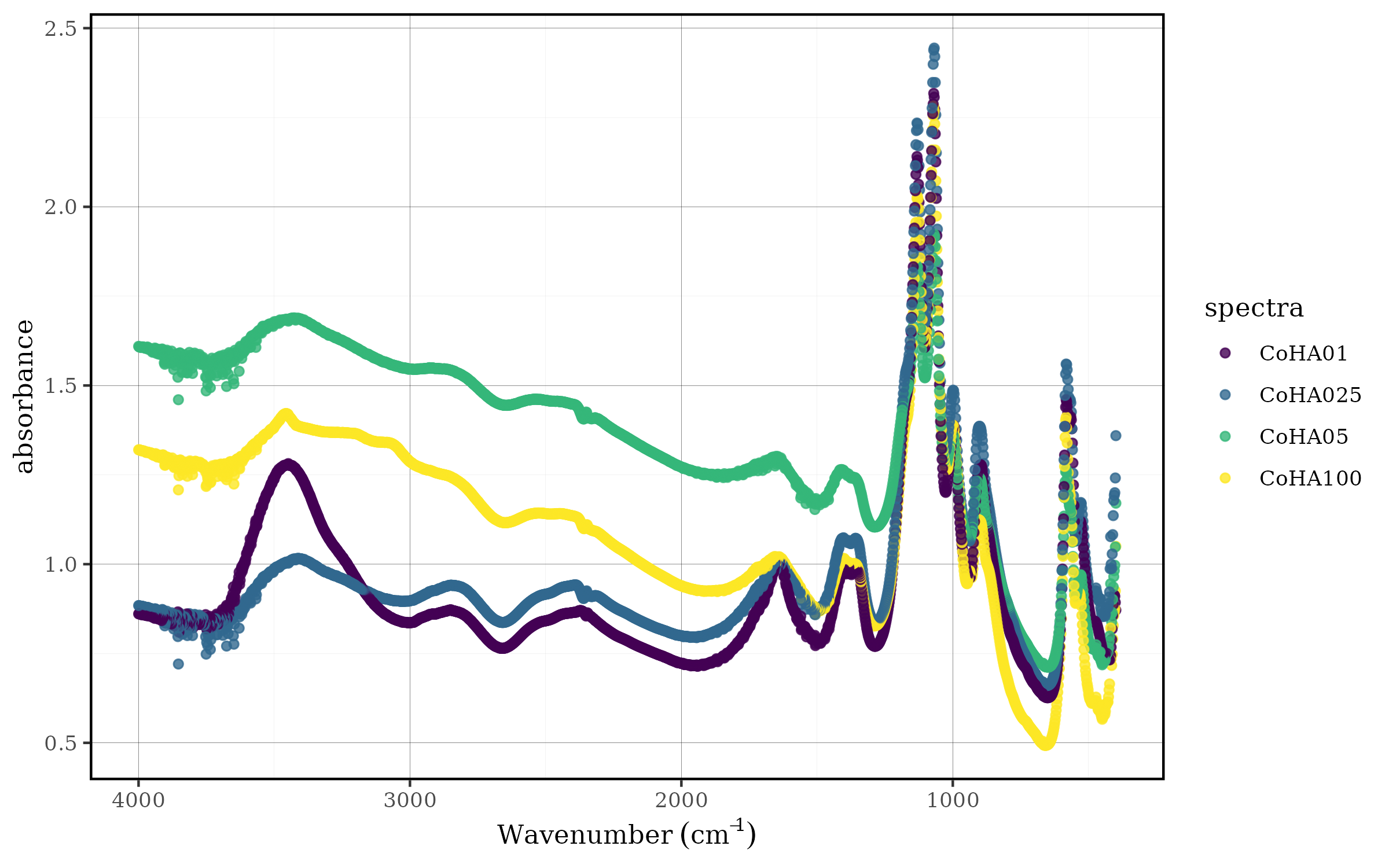
Select spectra
Tidyspec is designed to be easily integrated with the
entire tidyverse ecosystem. Then the user can easily choose
which spectra to keep/discard using dplyr::select.
CoHAspec %>%
dplyr::select(Wavenumber,CoHA100) %>%
tidyspec::spec_smartplot()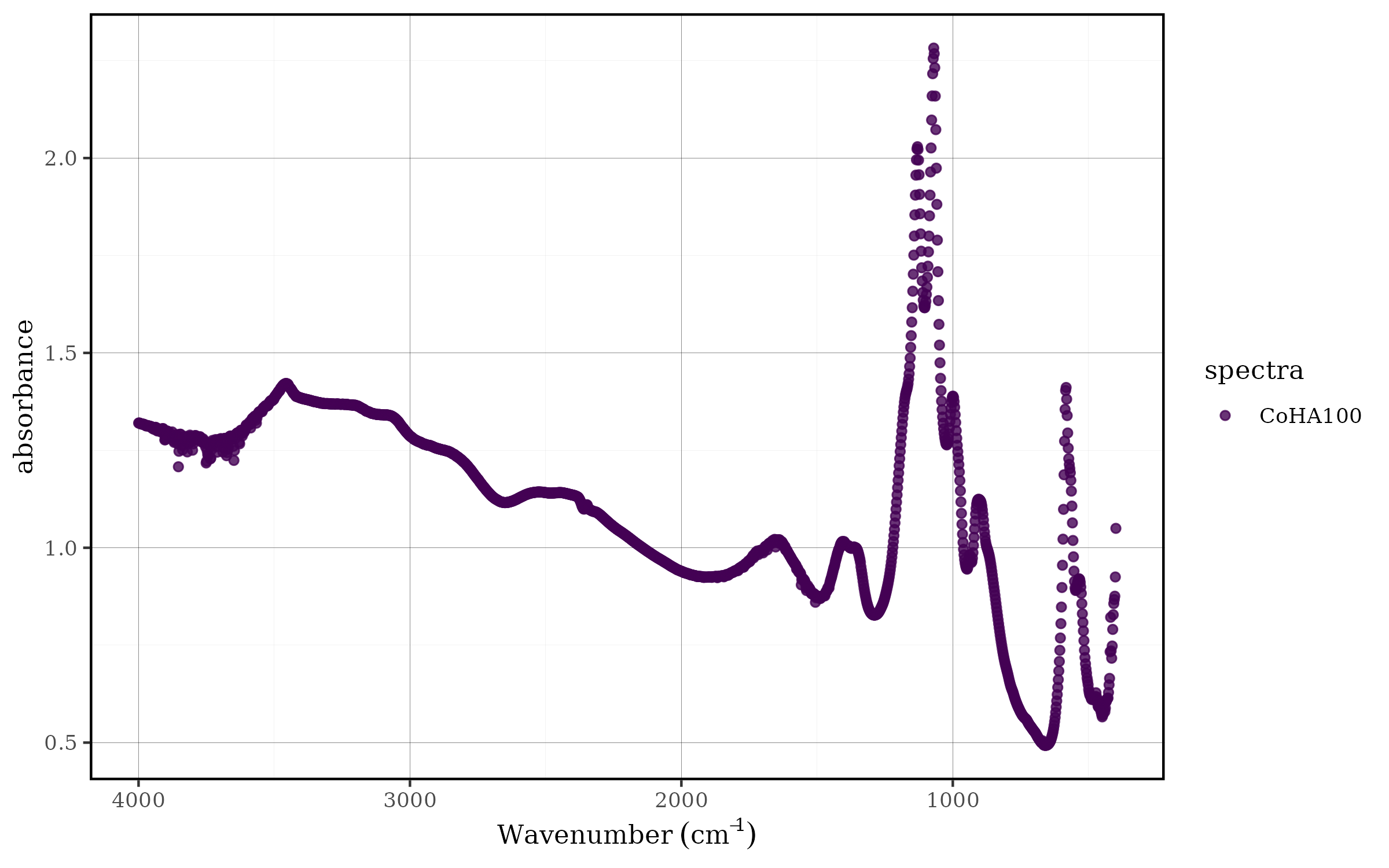
However, since tidyspec needs the column containing the
wave numbers and the user could end up messing up and not keeping it
during operations, we decided to develop spec_select which
simplifies the process.
CoHAspec %>%
tidyspec::spec_select(CoHA100) %>%
tidyspec::spec_smartplot()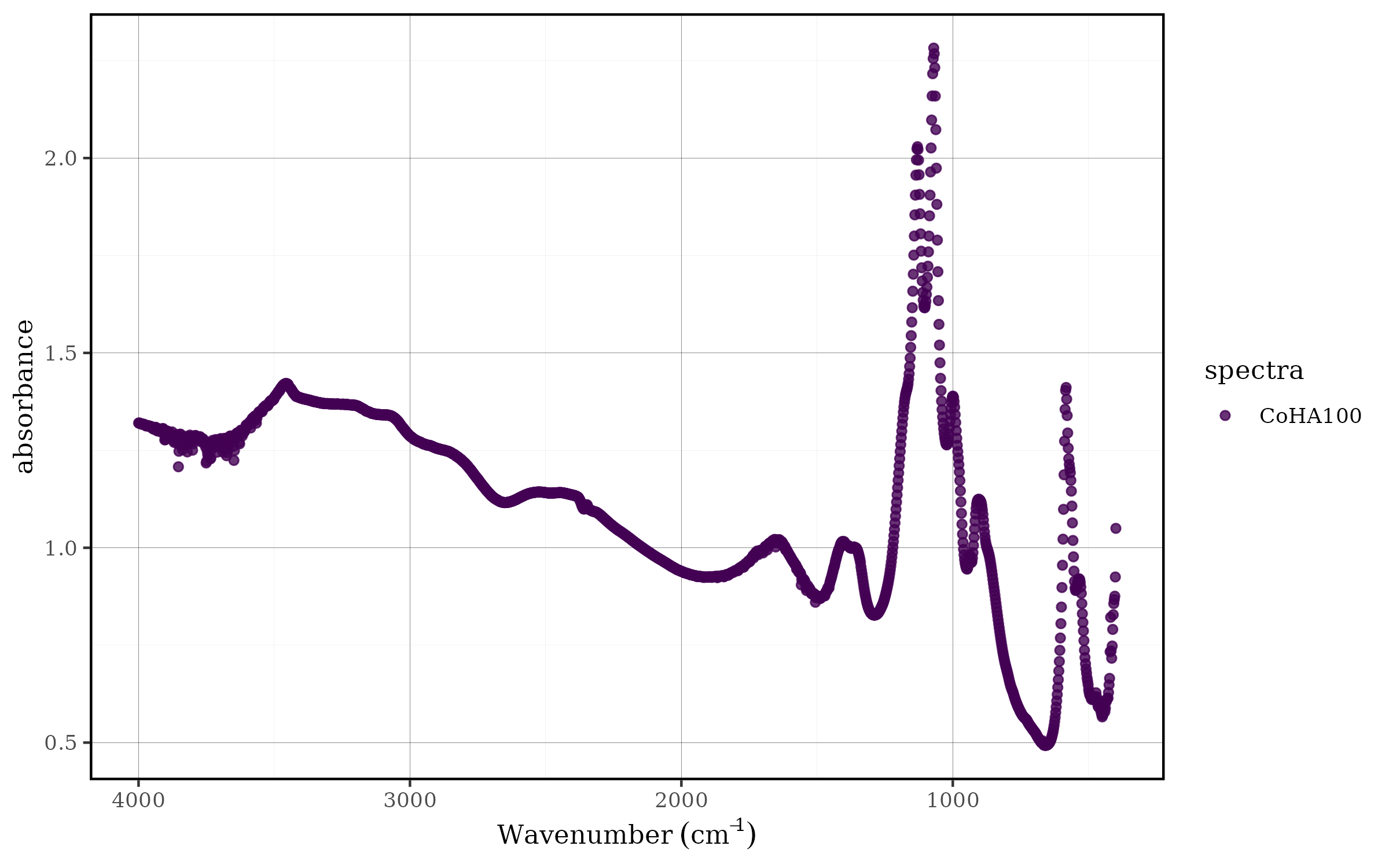
Filter spectra
Similarly to the spectrum selection process, filtering regions of the
spectrum can be done using dplyr::filter.
CoHAspec_filt <-
CoHAspec %>%
tidyspec::spec_select(CoHA100) %>%
dplyr::filter(Wavenumber > 1000,
Wavenumber < 1950)spec_filter simplifies the process of filtering the
desired regions.
CoHAspec_filt <-
CoHAspec %>%
spec_select(CoHA100) %>%
spec_filter(wn_min = 1000,
wn_max = 1950)
spec_smartplot(CoHAspec_filt, geom = "line")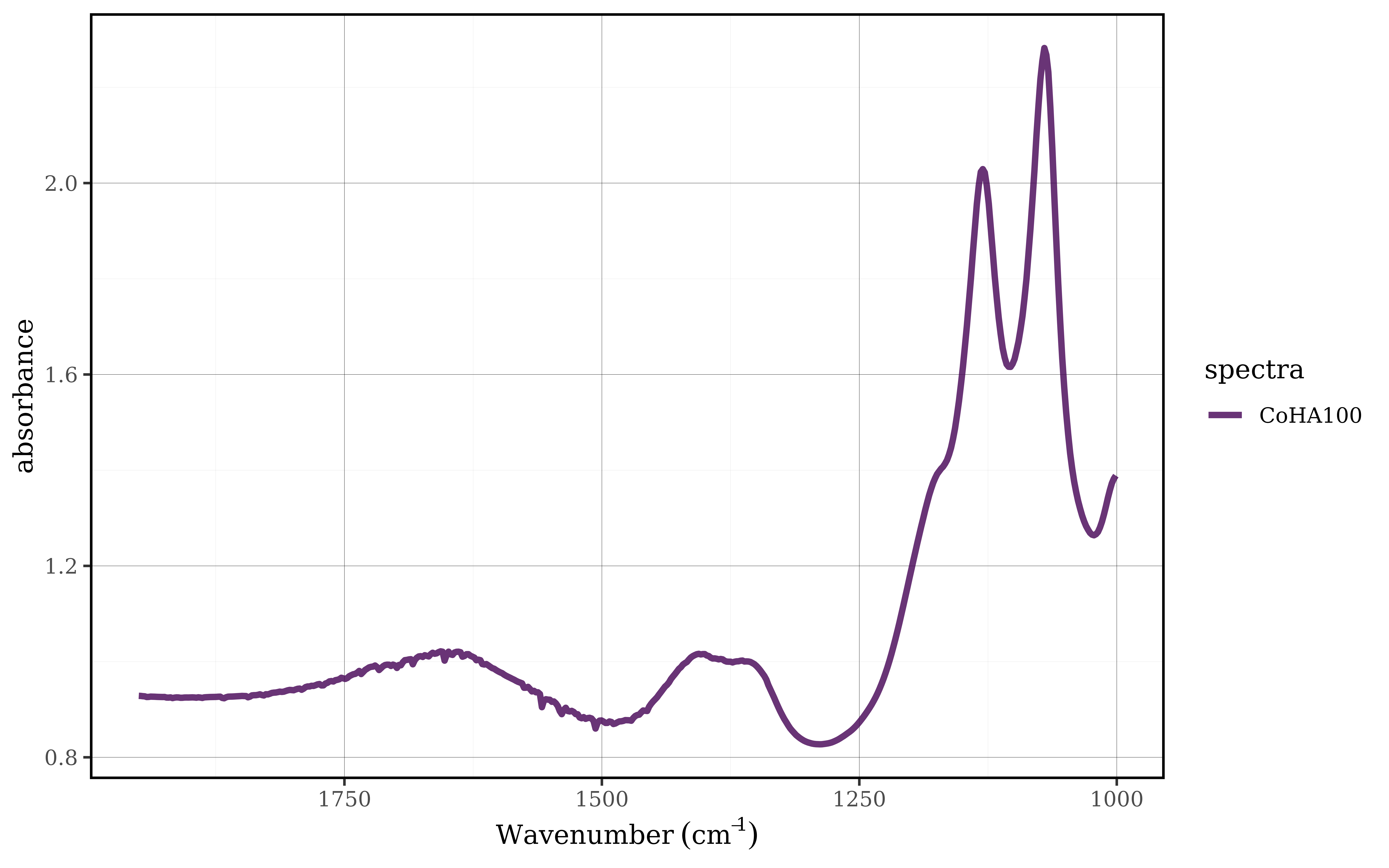
Smoothing the data
Spectroscopic data often contains noise that can interfere with
analysis and interpretation. Smoothing techniques help reduce this noise
while preserving the essential spectral features. The
tidyspec package provides two main smoothing methods:
moving averages and Savitzky-Golay filtering.
Moving averages method
The moving average method (spec_smooth_avg()) is one of
the simplest and most intuitive smoothing techniques. It works by
replacing each data point with the average of its neighboring points
within a specified window size. This method is particularly effective
for reducing random noise while maintaining the overall shape of
spectral peaks.
How it works:
- For each point, it calculates the mean of surrounding points within
a defined window
- Larger windows provide more smoothing but may blur important
features
- Smaller windows preserve detail but provide less noise reduction
Parameters:
-
wn_col: Column name for wavelength data (default: “Wn”)
-
window: Window size for the moving average (default: 15)
-
degree: Polynomial degree for smoothing (default: 2)
CoHAspec_filt %>%
spec_smooth_avg() %>%
spec_smartplot(geom = "line")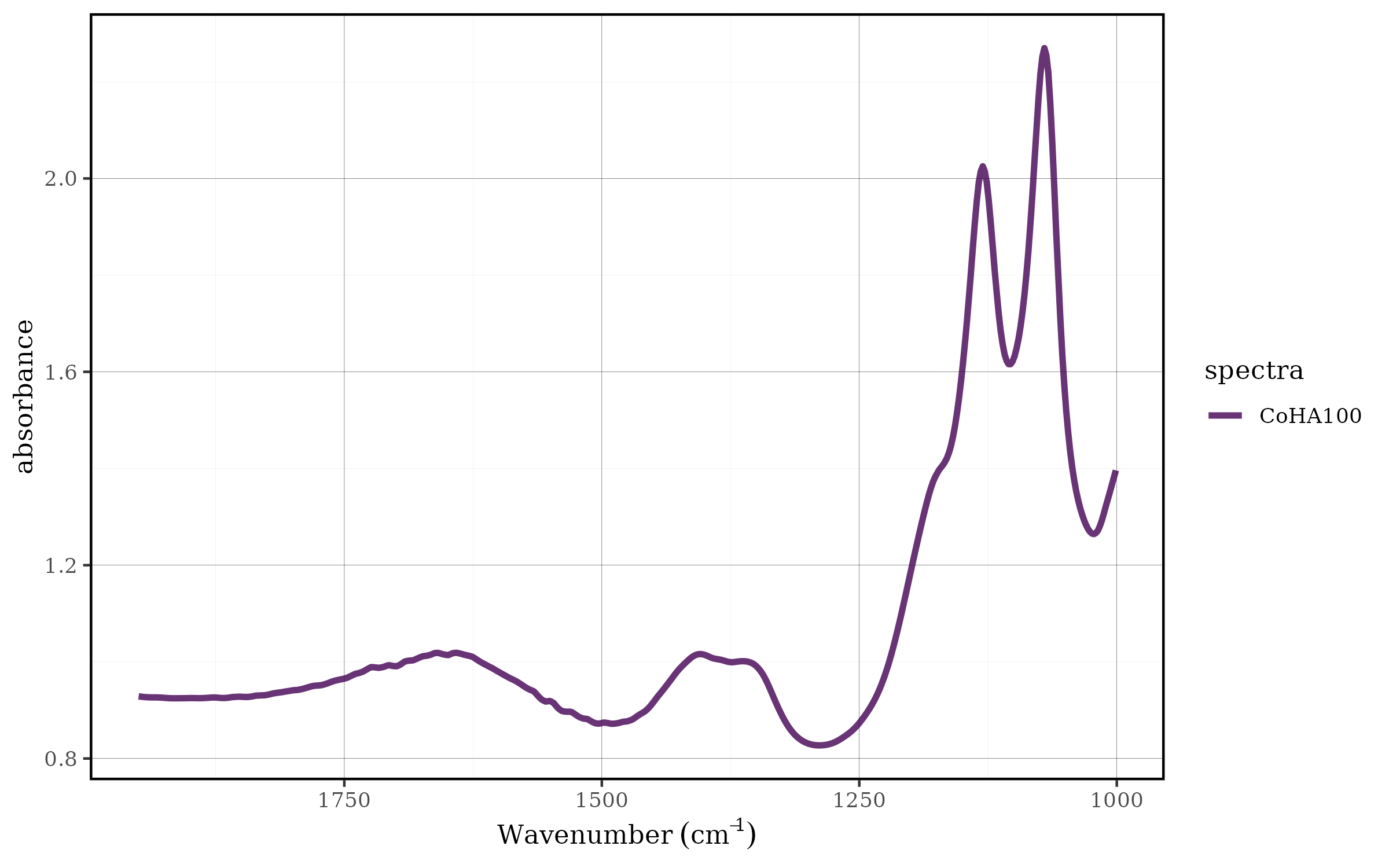
Savitz-Golay method
The Savitzky-Golay filter (spec_smooth_sga()) is a more
sophisticated smoothing technique that is particularly well-suited for
spectroscopic data. Unlike simple moving averages, this method fits a
polynomial to a local subset of data points and uses this polynomial to
estimate the smoothed value. This approach is especially effective at
preserving peak shapes, heights, and widths while reducing noise.
How it works:
- For each data point, it fits a polynomial of specified degree to
nearby points within a window
- The smoothed value is calculated from this local polynomial
fit
- This preserves the underlying spectral features better than simple
averaging
- The method can also calculate derivatives directly during the smoothing process
Parameters:
-
wn_col: Column name for wavelength data (default: “Wn”)
-
window: Window size for smoothing (default: 15, should be odd)
-
forder: Polynomial order for fitting (default: 4)
-
degree: Degree of differentiation (default: 0 = smoothing only)
CoHAspec_filt %>%
spec_smooth_sga() %>%
spec_smartplot(geom = "line")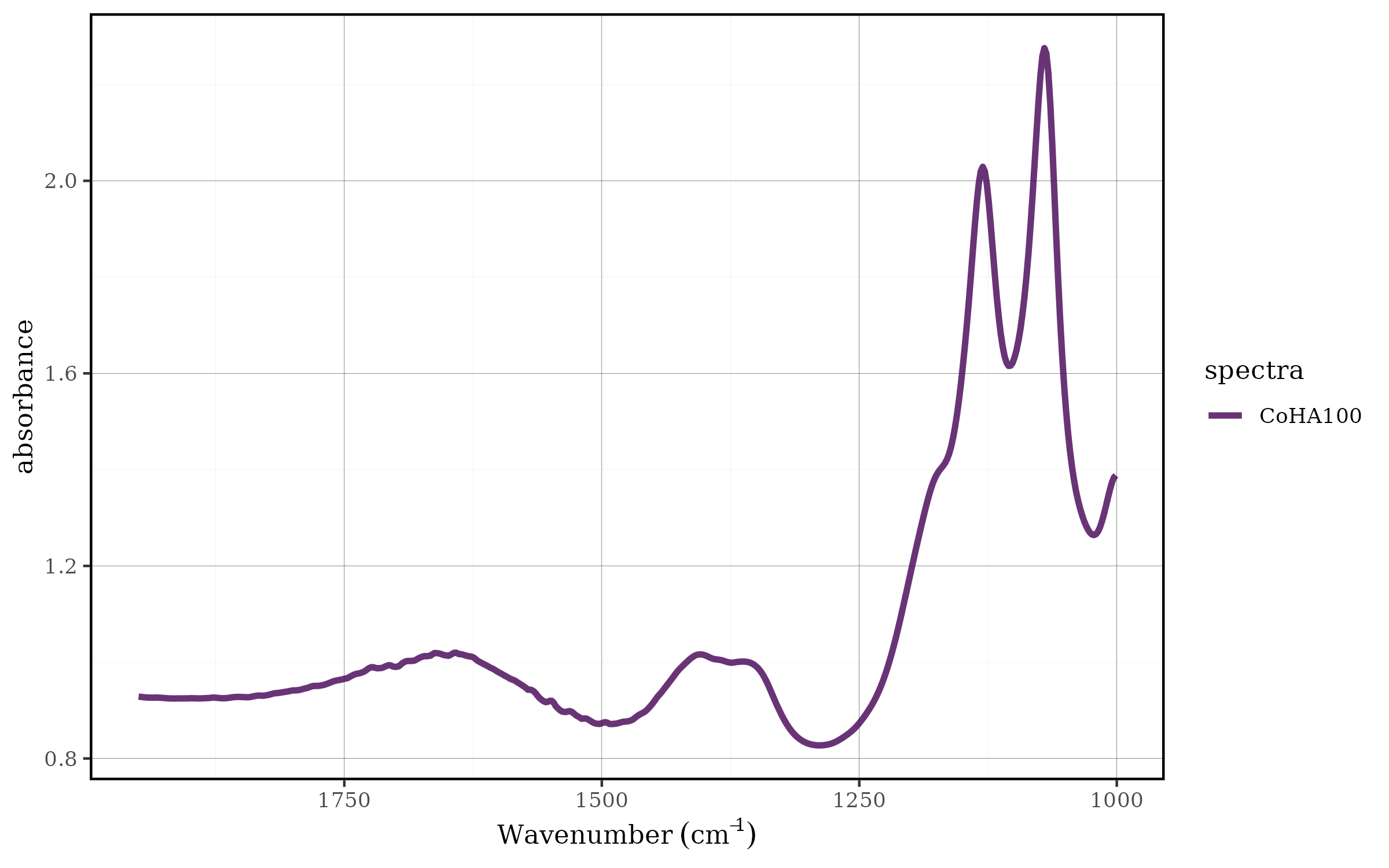
Derivatives
Differentiation of spectral data is a fundamental technique in
chemometrics that allows for highlighting specific spectral features,
reducing baseline effects, and improving the resolution of overlapping
peaks. The spec_diff() function implements numerical
differentiation for spectral data, offering flexibility to calculate
first-order or higher-order derivatives.
Using the spec_diff() Function
The spec_diff() function accepts the following
arguments:
-
.data: A data.frame or tibble containing spectral data -
wn_col: A character string specifying the column name for the wavelength data (default: “Wn”) -
degree: A numeric value specifying the degree of differentiation. If degree = 0, the original data is returned without any changes
Example:
CoHAspec_filt %>%
tidyspec::spec_smooth_sga() %>%
spec_diff(degree = 2) %>%
tidyspec::spec_smartplot(geom = "line")
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_line()`).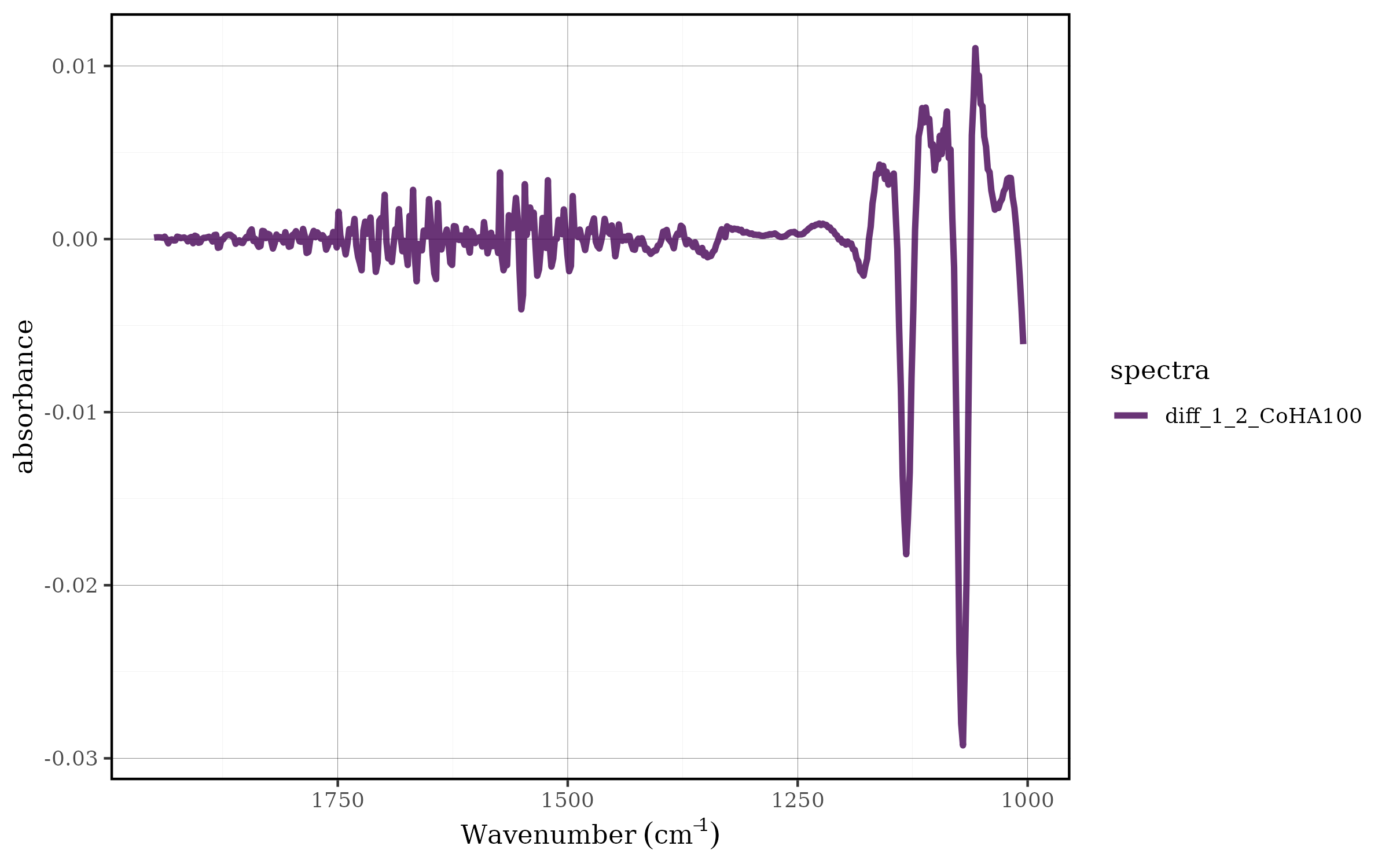
In this pipeline:
The filtered spectral data (CoHAspec_filt) is first smoothed using the Savitzky-Golay algorithm
Then, a second-order derivative is applied using
spec_diff(degree = 2)The result is visualized through
spec_smartplot()with line geometry
Baseline correction
Baseline correction is a crucial preprocessing step in spectral analysis that removes systematic variations in the baseline that are not related to the analyte of interest. These baseline variations can arise from instrumental drift, scattering effects, sample positioning, or optical interferences. Proper baseline correction enhances spectral quality and improves the accuracy of subsequent quantitative and qualitative analyses.
Rolling ball method
The rolling ball algorithm is a popular baseline correction technique that estimates the baseline by “rolling” a virtual ball of specified radius beneath the spectrum. This method is particularly effective for correcting baselines with smooth, curved variations and is widely used in various spectroscopic applications.
Function Parameters
The spec_blc_rollingBall() function provides
comprehensive control over the baseline correction process:
-
.data: A data.frame or tibble containing spectral data -
wn_col: Column name for wavelength data (default: “Wn”) -
wn_min: Minimum wavelength for baseline correction range -
wn_max: Maximum wavelength for baseline correction range -
wm: Window size of the rolling ball algorithm (controls ball radius) -
ws: Smoothing factor of the rolling ball algorithm -
is_abs: Logical indicating if data is in absorbance format
Baseline Correction Application
The following example demonstrates baseline correction applied to a specific wavelength range:
CoHAspec_filt %>%
spec_smooth_sga() %>%
spec_blc_rollingBall(wn_min = 1030,
wn_max = 1285,
ws = 10,
wm = 50) %>%
tidyspec::spec_smartplot(geom = "line")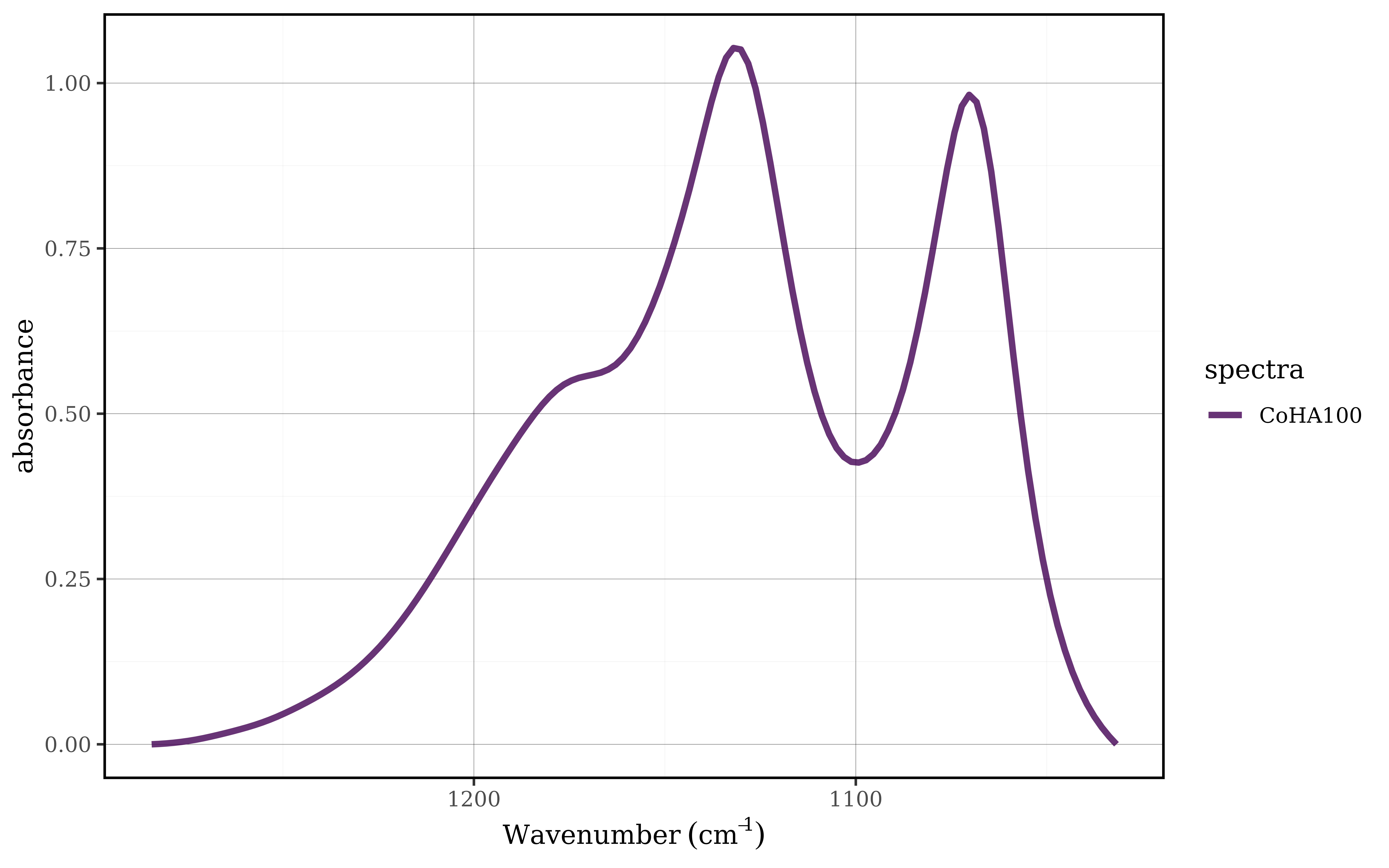
In this pipeline:
- Smoothing is applied first to reduce noise
- Rolling ball correction is performed in the 1030-1285 cm⁻¹
range
- Window size (wm = 50) and smoothing factor (ws = 10) are optimized for the data
Looking to the baseline
To better understand the correction process, we can examine the
estimated baseline using spec_bl_rollingBall():
CoHAspec_filt %>%
spec_smooth_sga() %>%
spec_bl_rollingBall(wn_col = "Wavenumber",
wn_min = 1030,
wn_max = 1285,
ws = 10,
wm = 50) %>%
spec_smartplot()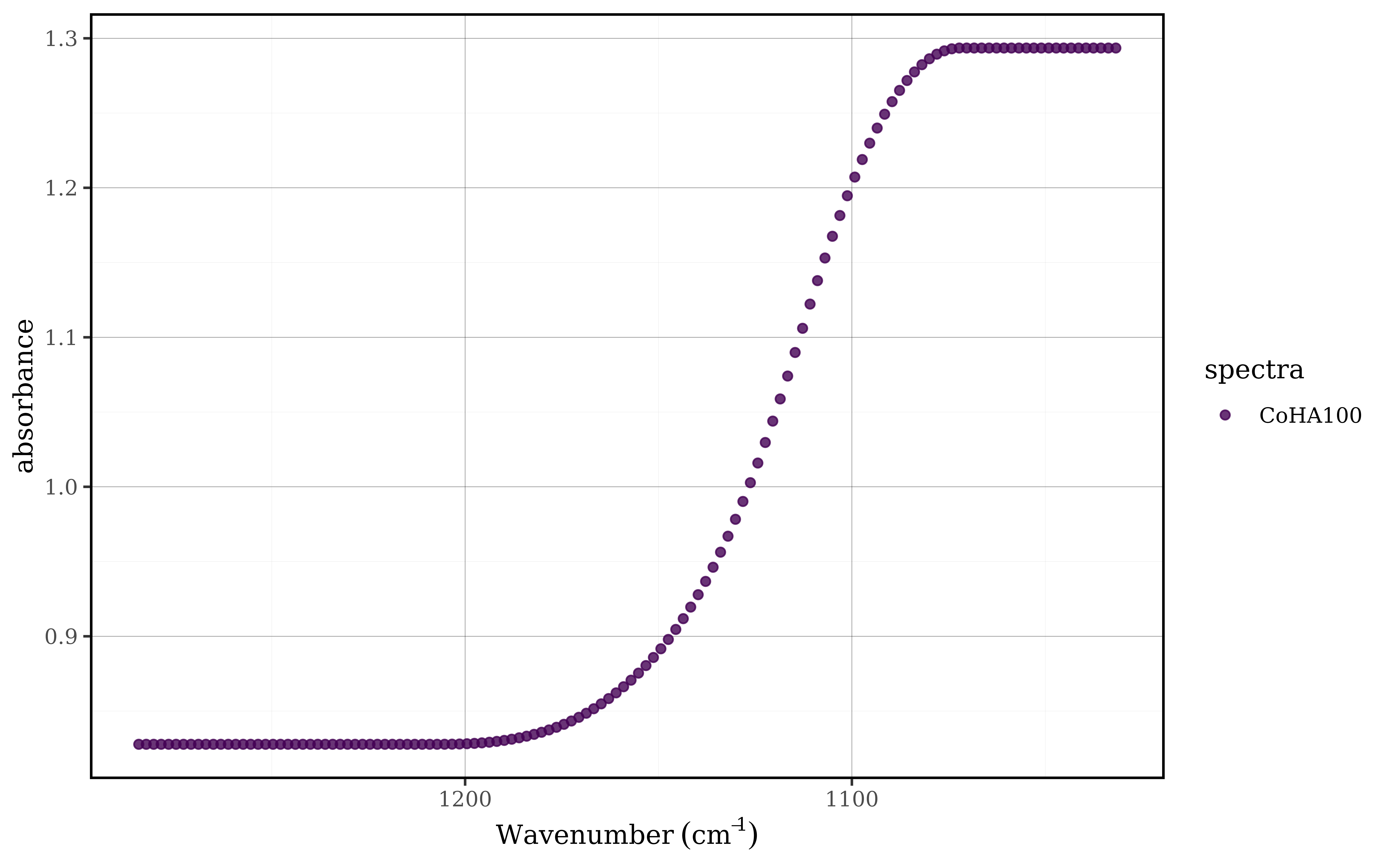
This visualization shows only the estimated baseline, allowing you to assess whether the algorithm properly captures the underlying baseline trend.
Comparing Original and Baseline
For a comprehensive view of the correction process, we can overlay the original spectrum with the estimated baseline:
bl <- CoHAspec_filt %>%
spec_smooth_sga() %>%
spec_bl_rollingBall(wn_col = "Wavenumber",
wn_min = 1030,
wn_max = 1285,
ws = 10, wm = 50)
CoHAspec_filt %>%
spec_smooth_sga() %>%
spec_filter(wn_min = 1030, wn_max = 1285) %>%
left_join(bl) %>%
spec_smartplot(geom = "line")
#> Joining with `by = join_by(Wavenumber, CoHA100)`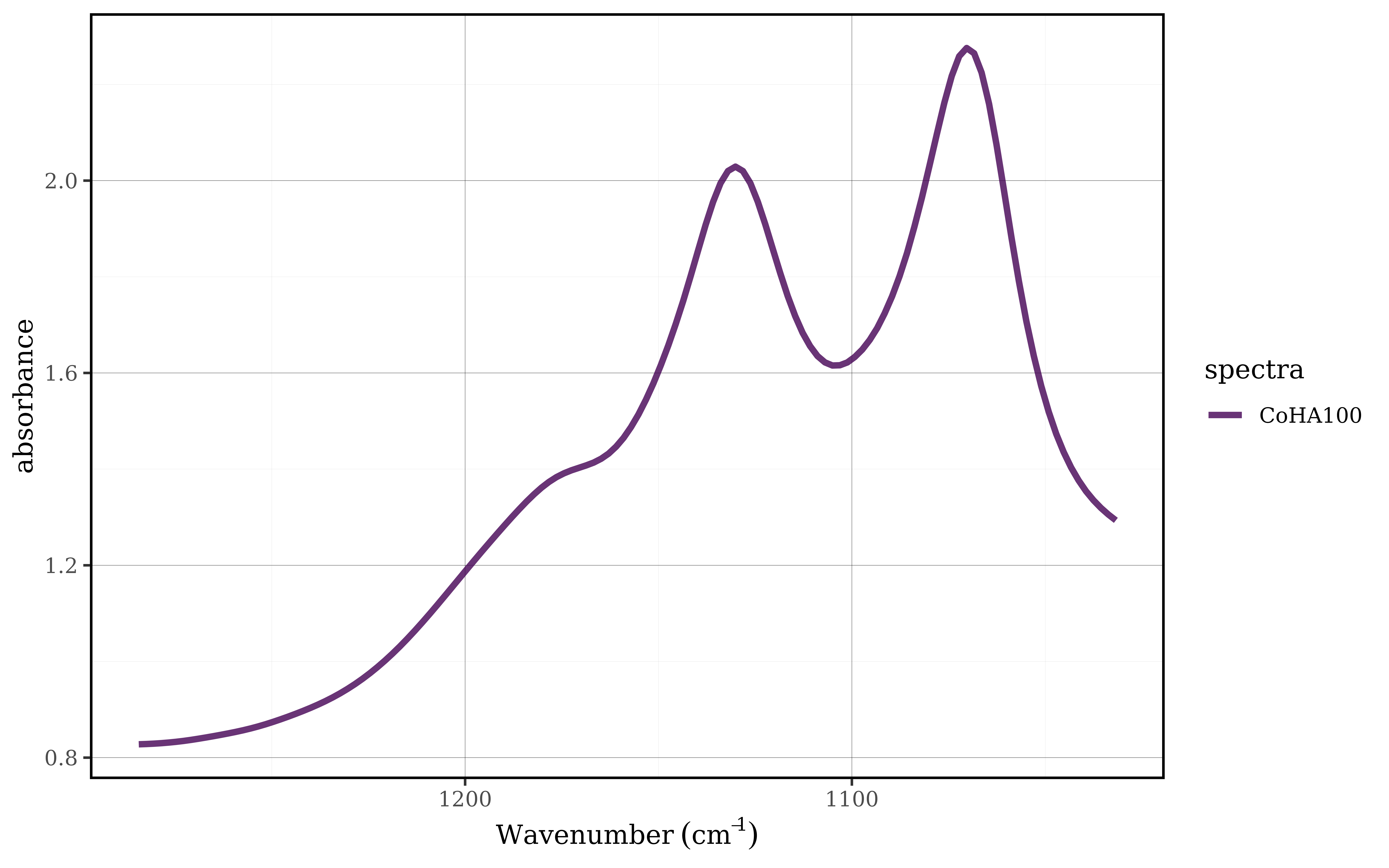
This comparison allows you to:
- Evaluate the baseline estimation quality
- Identify regions where the algorithm may need parameter adjustment
- Verify that the baseline follows the expected spectral trend
Scaling the spectra
Spectral scaling (normalization) is a preprocessing technique that standardizes spectral data to facilitate comparison between samples and improve the performance of multivariate analysis methods. Different scaling approaches address various sources of systematic variation in spectral data, such as differences in sample concentration, path length, or instrumental response.
Importance of Spectral Scaling
Scaling is essential when:
_ Sample concentrations vary: Different analyte concentrations can mask spectral patterns _ Path lengths differ: Variations in sample thickness affect overall intensity _ Instrumental variations exist: Different measurement conditions create systematic offsets _ Multivariate analysis is planned: Many chemometric methods assume standardized data _ Pattern recognition is the goal: Scaling emphasizes spectral shape over absolute intensity
Working with a Spectral Region
For demonstration purposes, we’ll work with a specific spectral region that contains relevant analytical information:
CoHAspec_region <- CoHAspec %>%
dplyr::filter(Wavenumber > 1300,
Wavenumber < 1950)
CoHAspec_region %>%
spec_smartplot(geom = "line")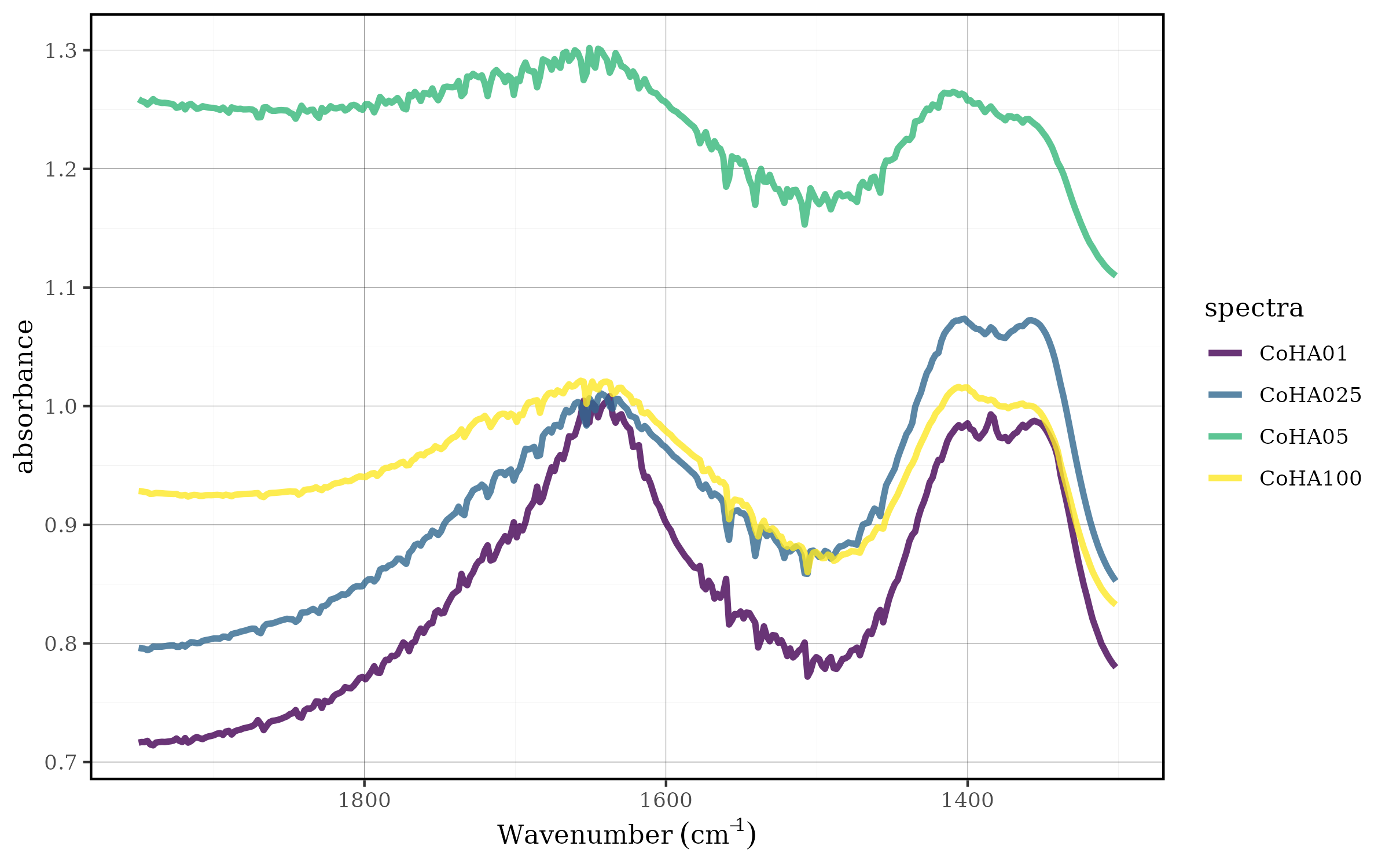
This region (1300-1950 cm⁻¹) is selected to focus on specific spectral features while avoiding regions that might contain noise or irrelevant information.
Min-Max Normalization (0-1 Scaling)
Min-max normalization scales each spectrum to a range between 0 and 1, preserving the relative relationships between spectral intensities while standardizing the dynamic range.
Mathematical Foundation
For each spectrum, the transformation is:
- Normalized value = (Value - Minimum) / (Maximum - Minimum)
- Results in values ranging from 0 to 1
- Preserves the original spectral shape
CoHAspec_region %>%
spec_norm_01() %>%
spec_smartplot(geom = "line")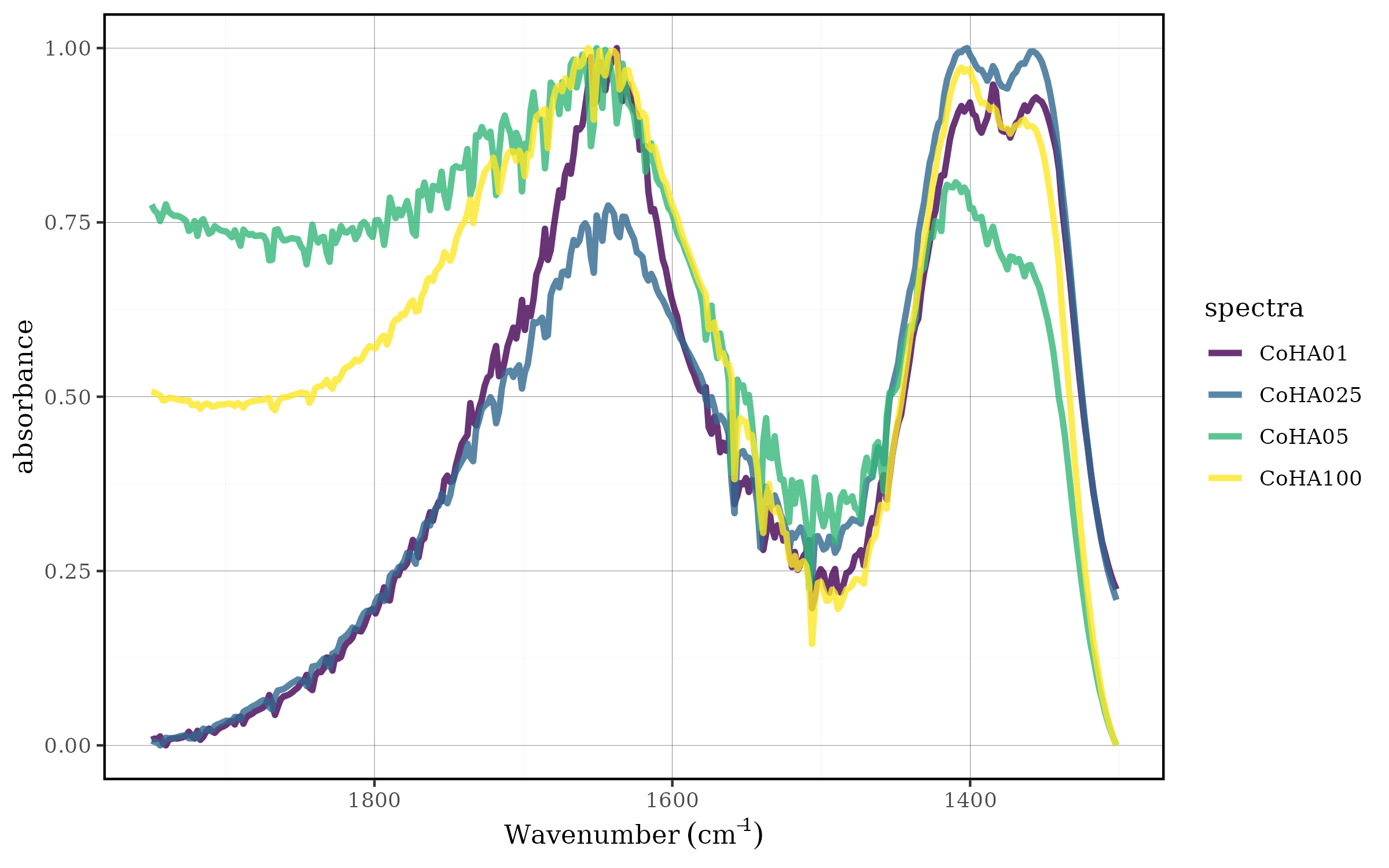
The spec_norm_01() function:
- Finds the minimum and maximum values for each spectrum
- Applies the min-max transformation
- Maintains spectral shape while standardizing intensity range
- Is particularly useful when absolute intensity differences are not analytically relevant
When to Use Min-Max Normalization
Min-max scaling is ideal when:
- You want to preserve spectral shape
- Absolute intensities vary due to concentration differences
- Visual comparison of spectral patterns is important
- The dynamic range needs standardization
The user can set any min and max value with the
spec_norm_minmax() function:
CoHAspec_region %>%
spec_norm_minmax(min = 1, max = 2) %>%
spec_smartplot(geom = "line")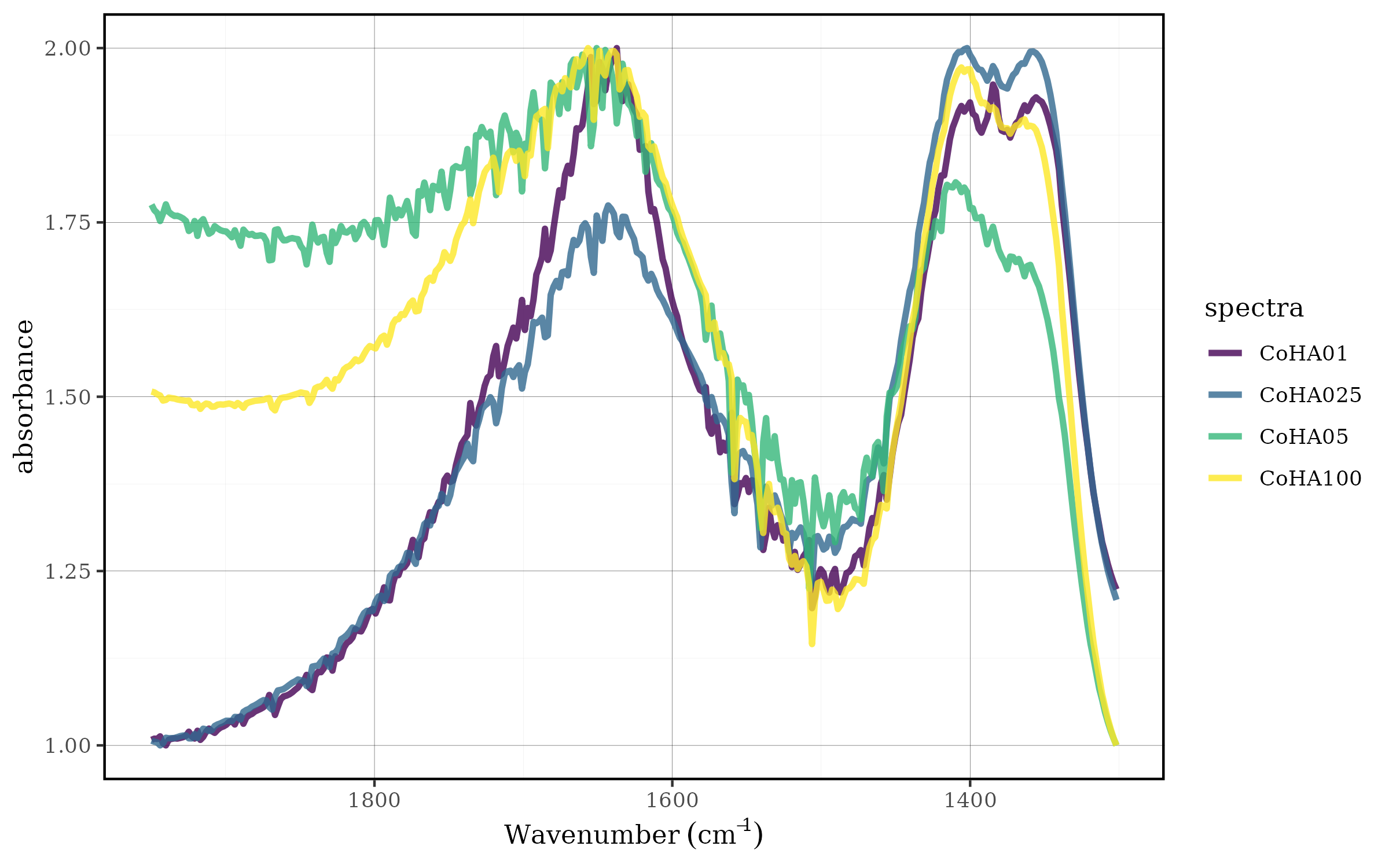
Standardization (Z-score Normalization)
Standardization transforms each spectrum to have a mean of 0 and standard deviation of 1, emphasizing deviations from the average spectral response.
Mathematical Foundation
For each spectrum, the transformation is:
- Standardized value = (Value - Mean) / Standard Deviation
- Results in mean = 0 and variance = 1
- Emphasizes relative variations around the mean
Application
CoHAspec_region %>%
spec_norm_var() %>%
spec_smartplot(geom = "line")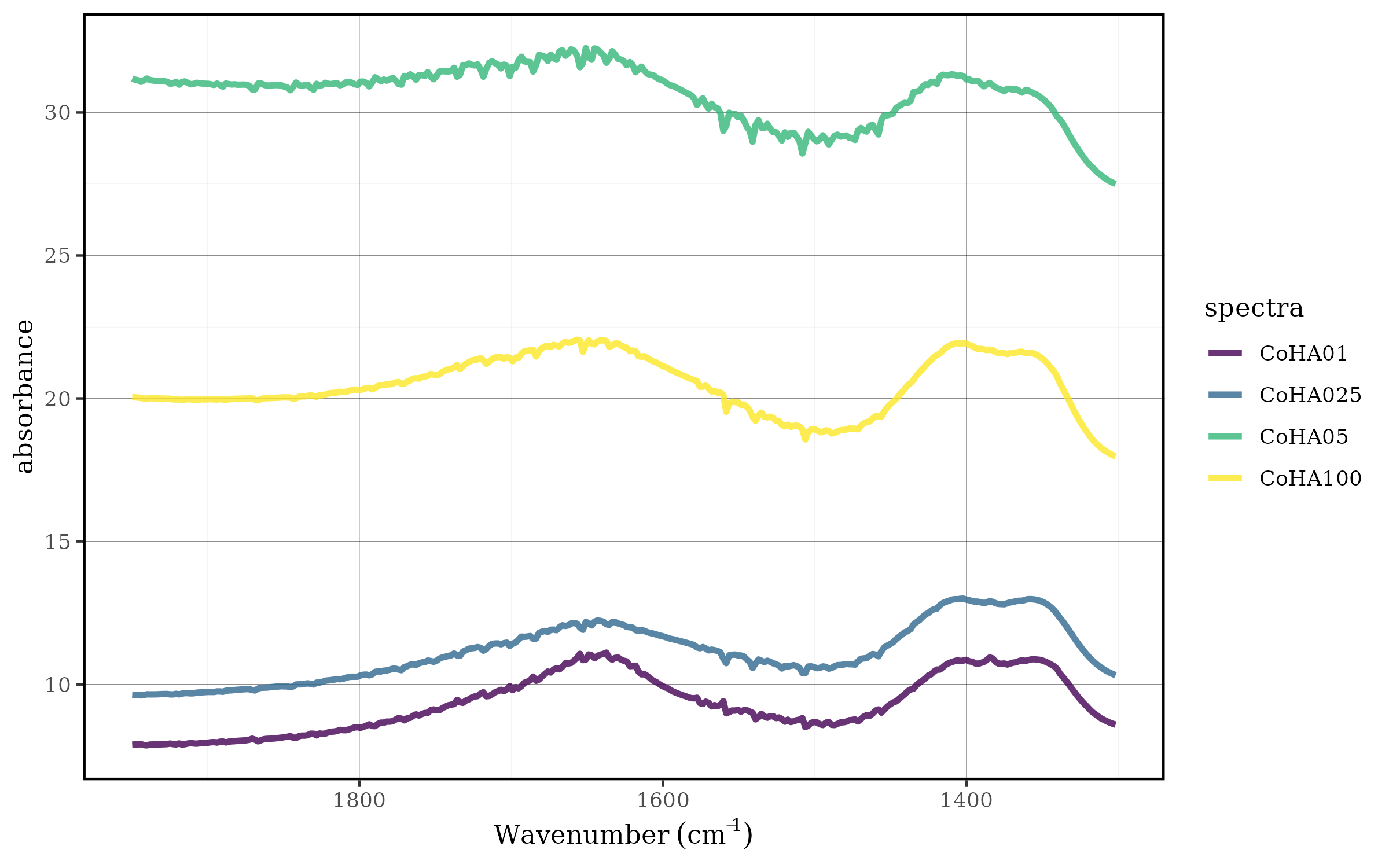
The spec_norm_var() function:
- Calculates the mean and standard deviation for each spectrum
- Applies z-score transformation
- Centers the data around zero
- Standardizes the variance across all spectra
Choosing the Right Scaling Method
The choice between scaling methods depends on your analytical objectives:
| Method | Best for | Preserves | Emphasizes |
|---|---|---|---|
| Min-Max (0-1) | Pattern recognition, visual comparison | Spectral shape | Relative intensities |
| Standardization | Multivariate analysis, variance-based methods | Spectral variations | Deviations from mean |
Integration with tidyverse
One of the key strengths of tidyspec is its seamless
integration with the tidyverse ecosystem. This design philosophy allows
users to leverage the full power of dplyr,
tidyr, purrr, and other tidyverse packages
to create sophisticated spectral analysis workflows. The following
examples demonstrate how to combine tidyspec functions with
tidyverse tools to solve common analytical challenges.
Case 1: Choosing the best baseline parameters
Parameter optimization is a critical aspect of spectral
preprocessing. The rolling ball baseline correction method has two key
parameters: ws (smoothing factor) and wm
(window size), and finding the optimal combination often requires
systematic evaluation across multiple parameter sets. The tidyverse
approach makes this process both elegant and efficient.
Setting up the parameter grid
First, we prepare our spectral data by focusing on a specific region and applying initial smoothing:
dados_1030_1285 <- CoHAspec_filt %>%
tidyspec::spec_smooth_sga() %>%
dplyr::filter(Wavenumber <= 1285, Wavenumber >= 1030)Next, we create a comprehensive parameter grid using
tidyr::crossing(), which generates all possible
combinations of the specified parameter values:
params <- tidyr::crossing(ws_val = c(2,4,6,8,10,12),
wm_val = c(10, 25, 40, 50))
params
#> # A tibble: 24 × 2
#> ws_val wm_val
#> <dbl> <dbl>
#> 1 2 10
#> 2 2 25
#> 3 2 40
#> 4 2 50
#> 5 4 10
#> 6 4 25
#> 7 4 40
#> 8 4 50
#> 9 6 10
#> 10 6 25
#> # ℹ 14 more rowsThis approach creates a systematic grid of 24 parameter combinations (6 × 4), ensuring thorough exploration of the parameter space.
Applying baseline correction across parameters
The power of the tidyverse becomes evident when we need to apply the
same operation across multiple parameter sets. Using
dplyr::mutate() with purrr::pmap(), we can
efficiently apply baseline correction to each parameter combination:
df <- params %>%
dplyr::mutate(spectra = list(dados_1030_1285)) %>%
dplyr::mutate(spectra_blc = purrr::pmap(list(spectra, ws_val, wm_val),
\(x, y, z ) spec_blc_rollingBall(.data = x, ws = y, wm = z))) %>%
dplyr::mutate(title = paste0("ws = ", ws_val," , wm = ", wm_val )) %>%
dplyr::mutate(plot = purrr::map2(spectra_blc, title, ~ spec_smartplot(
.data = .x,
wn_col = "Wavenumber",
geom = "line") + labs(title = .y)
))This pipeline demonstrates several key tidyverse concepts:
- Nested data structures: Each row contains the complete spectral dataset
- Functional programming: purrr::pmap() applies the baseline correction function across multiple arguments
- Pipeline operations: Each step builds upon the previous one using the pipe operator
- Automated visualization: purrr::map2() creates customized plots for each parameter combination
Visualizing parameter optimization results
The final step involves creating comparative visualizations to assess the effect of different parameter combinations:
library(gridExtra)
#>
#> Attaching package: 'gridExtra'
#> The following object is masked from 'package:dplyr':
#>
#> combine
grid.arrange(grobs = df$plot[1:4], nrow = 2, ncol = 2)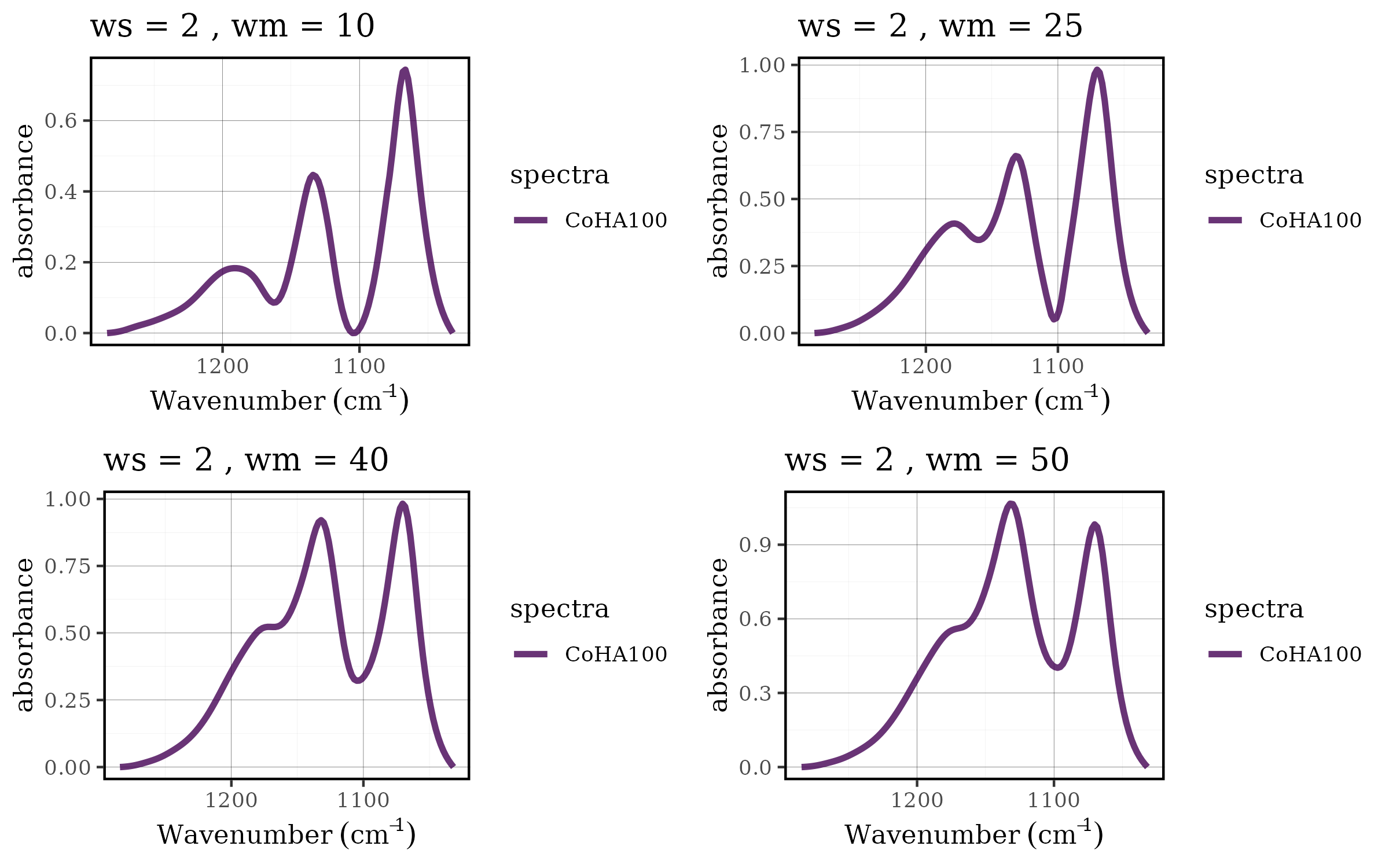
grid.arrange(grobs = df$plot[5:8], nrow = 2, ncol = 2)
grid.arrange(grobs = df$plot[9:12], nrow = 2, ncol = 2)
grid.arrange(grobs = df$plot[13:16], nrow = 2, ncol = 2)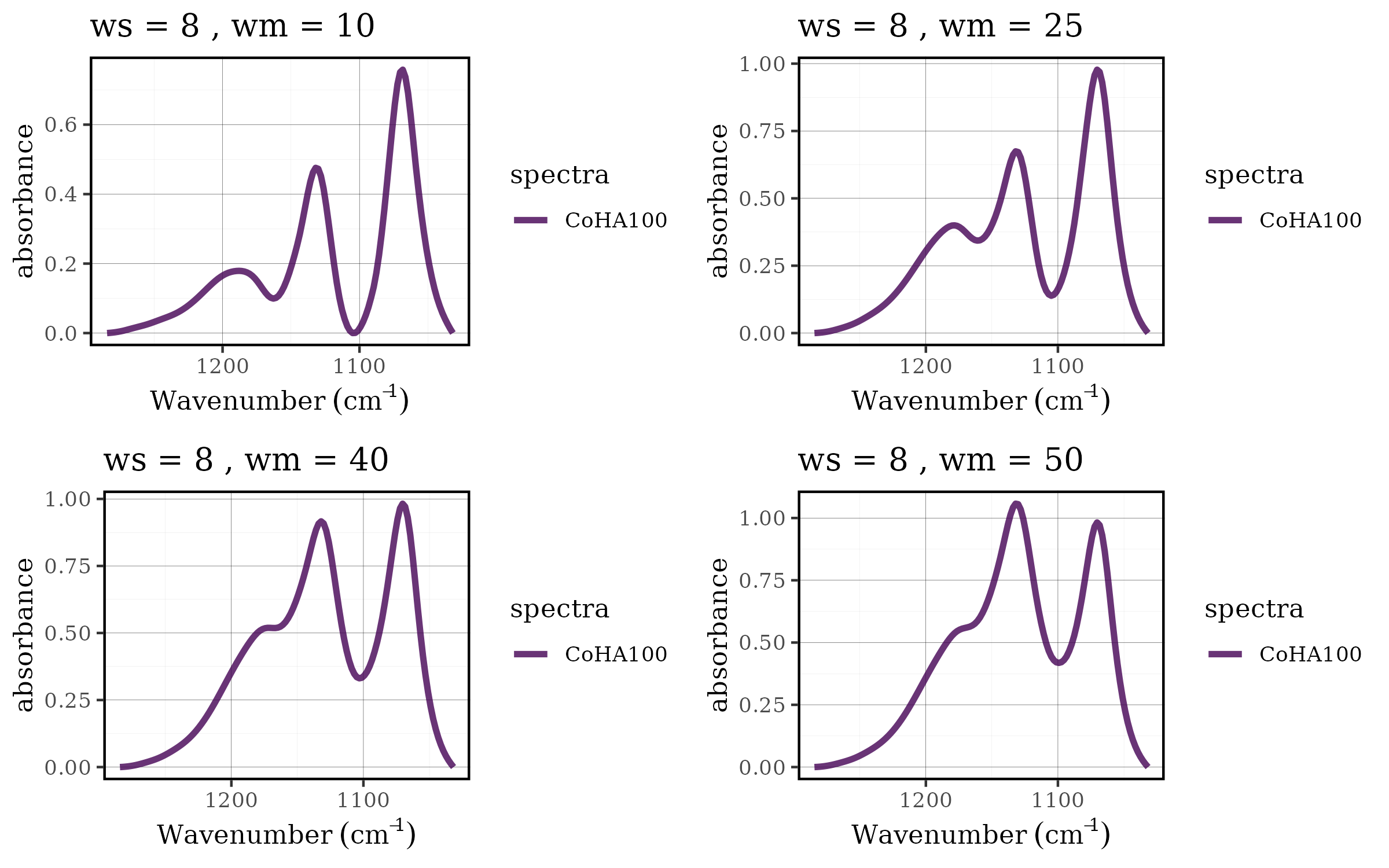
grid.arrange(grobs = df$plot[17:20], nrow = 2, ncol = 2)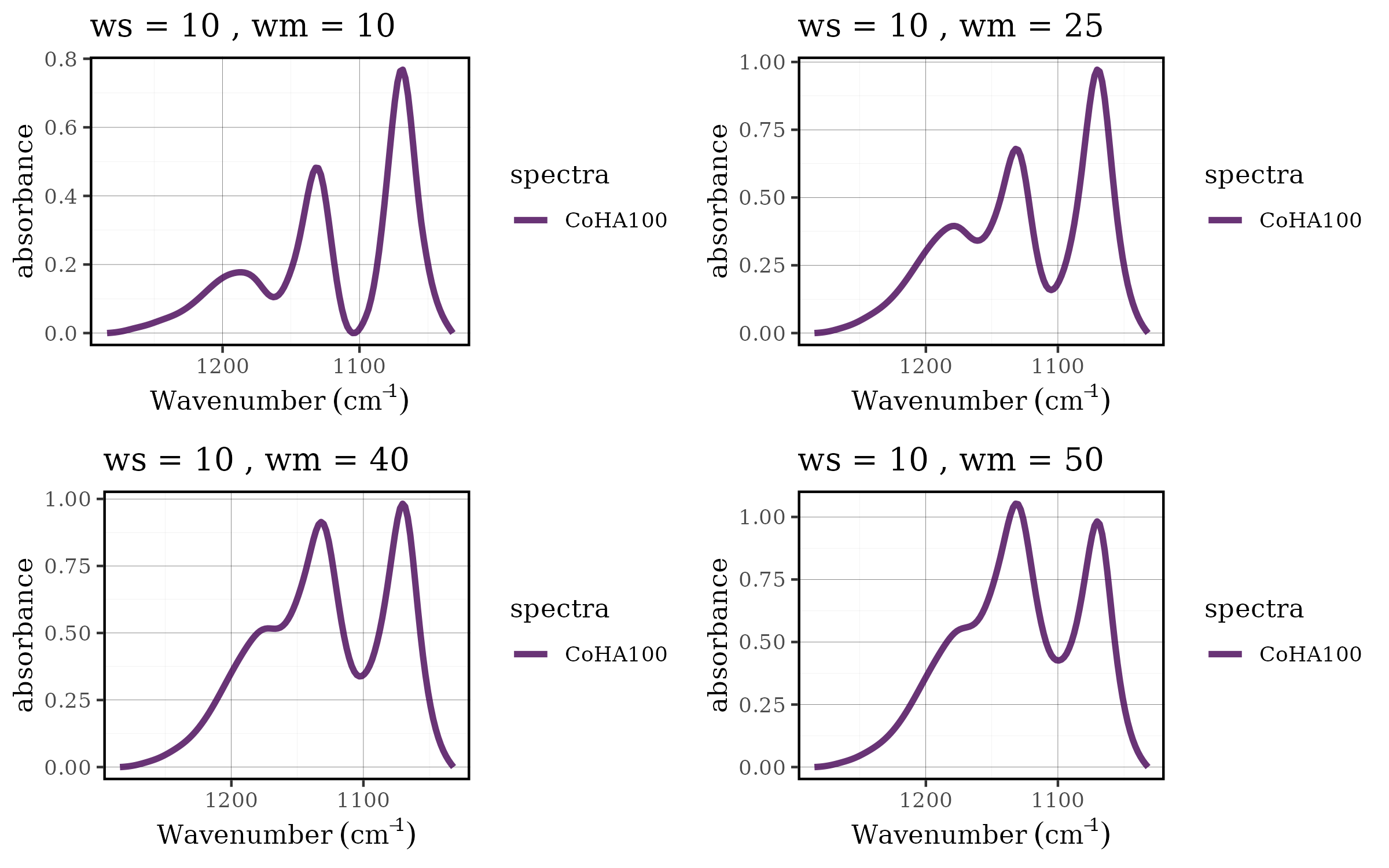
grid.arrange(grobs = df$plot[21:24], nrow = 2, ncol = 2)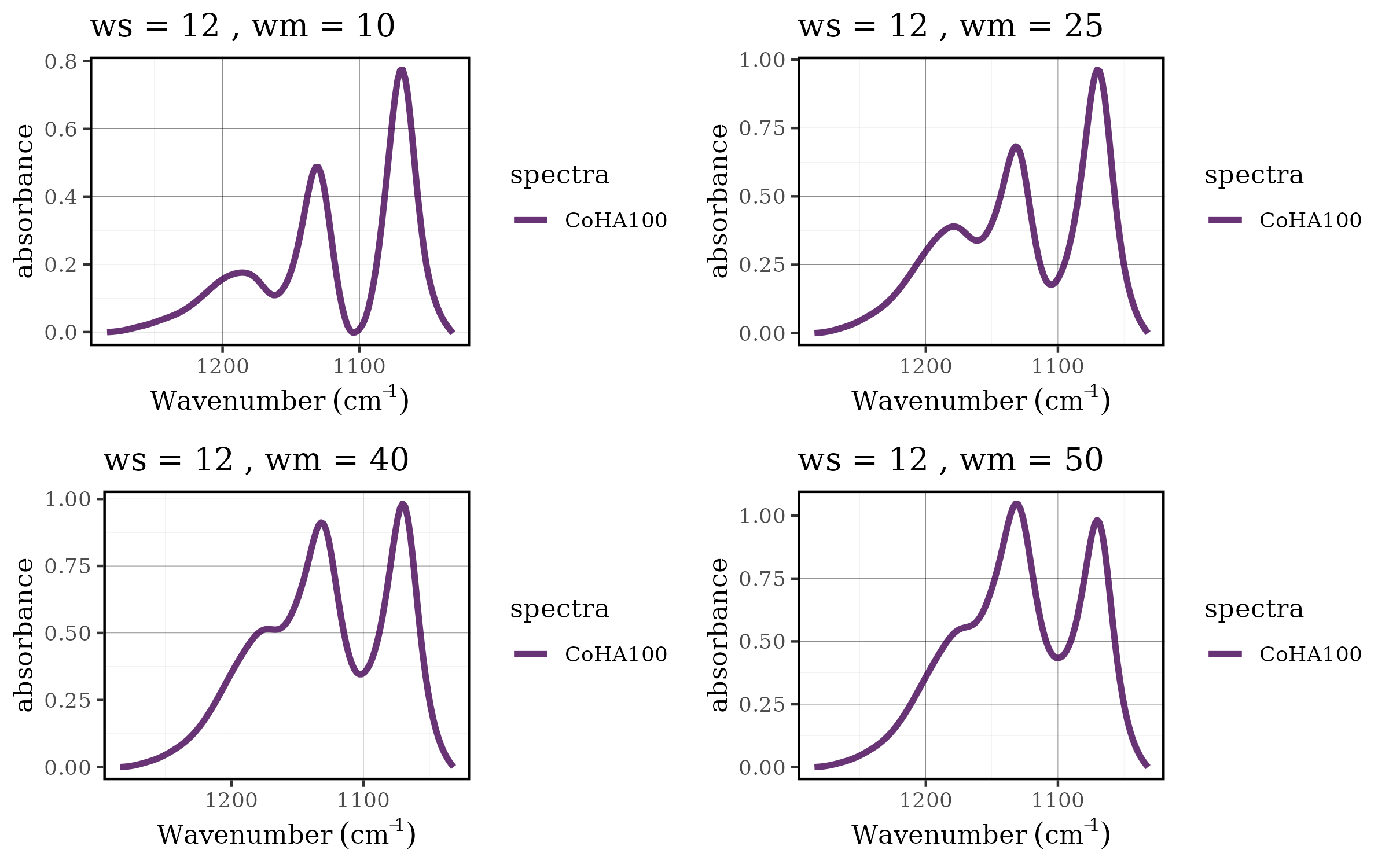
Benefits of the tidyverse approach
This workflow demonstrates several advantages of integrating tidyspec with the tidyverse:
- Scalability: Easy to add more parameters or parameter values
- Reproducibility: All steps are clearly documented and can be easily modified
- Efficiency: Vectorized operations handle multiple datasets simultaneously
- Flexibility: Each step can be customized or extended as needed
- Readability: The pipeline structure makes the analysis workflow transparent
Extending the approach
This methodology can be extended to optimize other spectral processing parameters:
- Smoothing parameters (window size, polynomial order)
- Normalization methods and ranges
- Derivative calculation parameters Spectral filtering ranges
The combination of tidyspec functions with tidyverse tools creates a powerful framework for systematic spectral analysis, enabling researchers to make data-driven decisions about preprocessing parameters while maintaining clean, reproducible code.
